UMaine researchers develop new model for predicting superspreader events in an epidemic
Two researchers with the School of Biology and Ecology at the University of Maine have developed a new model for quantifying the potential for the spread of disease across diverse environments and among varied population densities during an epidemic.
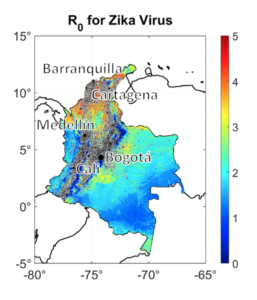
UMaine research associate Brandon Lieberthal and assistant professor Allison Gardner developed a mathematical analysis that simultaneously considers two probability-dependent epidemiologic factors to identify superspreader nodes that pose a high risk of propagating disease.
Previous predictive models evaluated connectivity of a location, or node, in a human mobility network and the disease reproduction rate, or R value, of a location independently. This study demonstrates that considered together, high network connectivity or a high infection rate create moderate-risk locations, while ratings of both factors above a certain threshold are required to designate a node as high-risk.
The study, “Connectivity, Reproduction Number, and Mobility Interact to Determine Communities’ Epidemiological Superspreader Potential in a Metapopulation Network,” was published in PLOS Computational Biology. The researchers expect that the new methodology can be applied in the future to compare and predict superspreader events and the efficacy of infection mitigation strategies in the context of disease systems with varied transmission modes, such as mosquito-borne viruses and SARS-CoV-2.
Contact: Joan Perkins, joan.perkins@maine.edu
