Overview
The Evolutionary Applications, or ‘EvoApps‘, Lab is the research group of Dr. Michael Kinnison at the University of Maine. The lab conducts basic and applied research at the broad interface of evolution, ecology and human interactions. Although popular perceptions of evolution bring to mind a ponderously slow and ancient process, that view has scientifically faded in favor of one where evolution is recognized as a dynamic and contemporary process that has implications for the fate and function of biodiversity in a rapidly changing world. Indeed, we contributed several of the field studies, experiments and meta-analysis that helped cement this new appreciation of contemporary evolution. Within this framework we consider genetic and phenotypic scales of diversity in wild or managed populations, with most projects investigating evolutionary or genetic signatures of human activities, or the role that ongoing evolution plays in shaping population, community and ecosystem conditions (eco-evolutionary dynamics). These themes are arguably important for our understanding and management of real-world challenges in fish and wildlife conservation, as well as for sustainable human health and food security. In addition to our evolutionary and genetic work, we also collaborate on projects related to other aspects of the conservation and ecology of aquatic species.
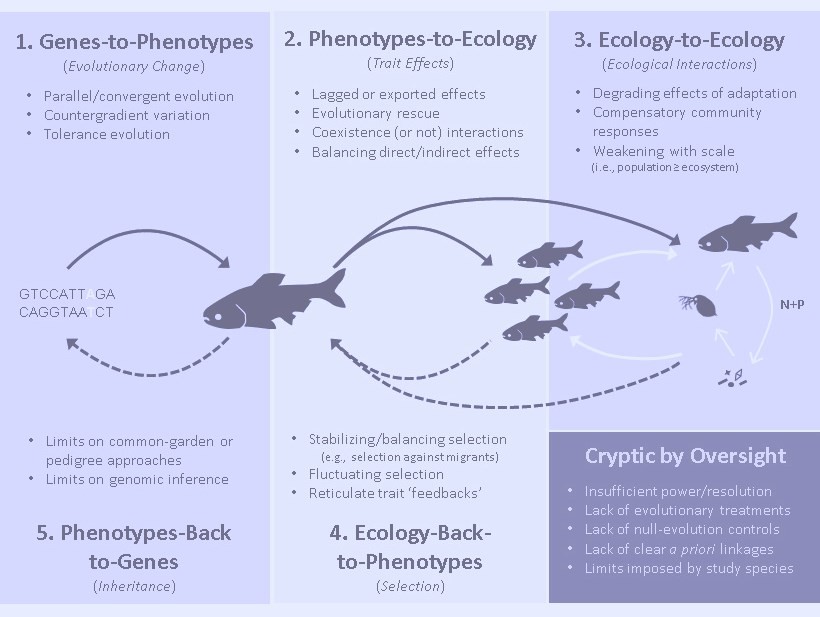
Eco-Evolutionary Dynamics and Contemporary Evolution
“It may be said that natural selection is daily and hourly scrutinising, throughout the world, every variation, even the slightest; rejecting that which is bad, preserving and adding up all that is good; silently and insensibly working, whenever and wherever opportunity offers, at the improvement of each organic being in relation to its organic and inorganic conditions of life. We see nothing of these slow changes in progress, until the hand of time has marked the long lapse of ages, and then so imperfect is our view into long past geological ages, that we only see that the forms of life are now different from what they formerly were.” – Charles Darwin, 1859.
In this one quote, Darwin captures what might be considered evolution’s dual-scales problem. On the one hand, we are reminded that the processes that drive evolution, such as natural selection, are operating all the time, and yet at the same time we are told the outcomes of those processes require geological time scales to become evident. In many ways, connecting these two time scales in a vivid fashion was one of Darwin’s greatest contributions, but the perception that evolution is vanishingly slow turns out to be vastly overstated.
Good examples of evolution occurring over humanly observable time spans began to appear, at least anecdotally, in the scientific literature as early as the late 1800s (e.g., industrial melanism in peppered moths). Throughout the 20th century dozens of other examples of such observable evolution were recognized and heralded as important exceptions that proved evolution’s rules. These examples were almost always labelled ‘rapid evolution’, under the presumption that any evolution that is observable must be exceptionally fast. However, by estimating and comparing rates from many studies, my colleague Andrew Hendry I showed that high rates of contemporary evolution are not really all that unusual. Indeed, they are the expectation where selection and other evolutionary processes are variable year-to-year and generation-to-generation. The slower rates measured over longer periods of time are due to temporal averaging of often substantial, but variable, short-term rates.
It might not seem a major leap, but recasting our perceptions of contemporary evolution from creeping slow, to frequently fast but chaotic, provides some impetus to reconsider evolution’s interactions with ecology. Certainly, it is hard to look at a detailed time series of variable trait change in nature and not suspect that dynamic ecological processes are at play shaping selectio. But the dynamic nature of contemporary evolution also raises the interesting opposite prospect, that we shouldn’t be too quick to dismiss contemporary evolution’s reciprocal role back on ecology. Although these reciprocal effects of evolution on ecology have attracted sporadic attention for decades, we are now in the midst of what some claim is the next great synthesis in ecology and evolution (Schoener 2011). My colleague Nelson Hairston Jr. and I referred to these reciprocal interactions of evolution and ecology as ‘eco-evolutionary dynamics’ in a 2007 paper, and the name caught on. The field has grown quite dramatically in the following decade and I encourage you to read Schoener’s Oxford Bibliography for a broader review of its development. The first book on ‘eco-evolutionary dynamics’ (by Andrew Hendry) is due out soon. Some recent and current projects on contemporary evolution and eco-evolutionary dynamics:
- Cryptic eco-evolutionary dynamics: A review of ‘cryptic eco-evolutionary dynamics’ with Nelson Hairston Jr. and Andrew Hendry that basically makes the case for how easy it can be to overlook such dynamics in nature and identifies the types of processes that can conceal the various links in an eco-evolutionary feedback.
- Eco-evolutionary trophic cascades: A collaborative NSF funded project on the potential role that evolution plays in determining the strength of trophic cascades in aquatic ecosystems. This work is being conducted using mosquitofish and in collaboration Dr. Eric Palkovacs.
- Plastic and transgenerational trait dynamics in mosquitofish: You can’t really study contemporary evolution or its contributions to ecology with simultaneously recognizing other processes shaping phenotypes within and among generations. Heather Arnett’s dissertation tackles these other sources of ecologically-relevant trait variation.
Anthropogenic and Managed Evolution
Humans have been called ‘the world’s greatest evolutionary force’ (Palumbi 2001). Although evolution doesn’t work via ‘forces’ per se (i.e., akin to physics), and one could argue whether other agents of selection are equal or greater (e.g., gravity), this perspective is probably pretty apt when it comes to encompassing the sheer scope of human effects on evolution in our modern world. Most work in this area, has focused on anthropogenic evolution in medical and agricultural settings. No doubt, the costs of human-induced evolution in those contexts, in terms of both money and human suffering, are immense. Not surprisingly, evolution in medical and agricultural systems is now heavily managed. It is not perfectly managed, but many practices and policies are in place for the express purpose of slowing unwanted evolution. The current state of affairs is quite different when we move beyond these completely anthropocentric contexts.
The fast generation times and large population sizes of many pathogens and pests, along with the harsh selection we impose through widespread use of chemical controls is a pretty tangible formula for evolutionary problems, but that should not be taken to imply such evolution is limited to some special organisms and circumstances. There is now growing appreciation that anthropogenic evolution is a much more expansive phenomenon. In a 2003 review paper, Craig Stockwell, Andrew Hendry and I made the case that many of the threats to conserving biodiversity, as well as the approaches we take to manage those threats, are potent drivers of contemporary evolution. In some cases, such evolution could be a valuable ally, but in other cases it could thwart even the best-intentioned efforts that ignore it. Since that early paper, I have been involved in a number of reviews and book chapters encouraging ways to better incorporate contemporary evolution in conservation, restoration and natural resource management. While some evolutionary challenges and solutions might be idiosyncratic to natural resource management, my colleagues and I tend to think that many challenges in food, health and the environment share commonalities that argue for a more broadly unified field of applied evolution (see Carroll et al. 2014). Indeed, we have suggested that evolutionary conservation biology and natural resource management could learn quite a lot from agriculture and medicine when it comes to actively managing evolution and incentivizing stakeholders to do so.
In addition to these reviews and syntheses, my lab has worked on a diversity of projects surrounding anthropogenic selection and evolution in conservation and natural resource contexts. These projects span analyses of evolution in introduced and invasive populations, harvested populations, threatened populations under supportive breeding, and populations facing habitat changes due to climate change and urbanization. Some recent and current projects on anthropogenic and managed evolution:
- Prescriptive evolution: In a recent review paper my colleagues and I suggest that the choice of whether to manage evolution in conservation biology is really a false dichotomy. In most cases, not managing evolution is really tantamount to managing it in a haphazard fashion. We discuss how evolution might be managed in a more prescriptive fashion that balances risks.
- Evolutionary rescue in complex landscapes: An individual-based model of evolution’s role in determining persistent or extinction of organisms distributed across landscapes experiencing complex spatial and temporal change, as might occur under combined influences of climate change and urbanization. This model is part of Jared Homola’s PhD dissertation.
- Speed of harvest evolution: In a first of its kind met-analysis (Darimont et al. 2009) my colleagues and I quantified how fast traits change in fish, wildlife and plant populations under selective harvest by humans. Rates of evolution due to harvest are over three times faster than in natural contexts and faster than most other anthropogenic drivers.

Landscape Genetics and Genomics
It’s almost hard to imagine the study of evolution in wild populations, or attempting real-world applications of evolutionary principles, without the insights afforded by modern population genetic tools. Likewise, emerging genomic resources are quickly changing how evolutionary biologists elucidate the heritable underpinnings of potentially adaptive phenotypes. The last couple decades have seen an explosion of scale in these tools, from population genetic studies that initially examined gene flow among a few populations, to landscape scale studies that map the genetic permeability of natural and anthropogenic landforms. Genomic studies grew from rare studies examining single candidate loci to modern genome scans comprising hundreds of thousands of sequencing reads to identify likely regions under selection. Indeed, while historically these two areas of genetic investigation were often conducted in isolation due to their different focus on largely neutral (population genetics) versus adaptive (genomic) components of genetic diversity, even those distinctions are starting to fade with new looking increasingly to tackle landscape scale studies of genomic variation.
In our lab population genetic and genomic tools are often applied to systems that have historically or currently face human pressures that could shape population connectivity, gene flow and adaption. This includes the genetic outcomes of recent introductions, urbanization, or population recovery. These contemporary contexts present challenges for traditional population genetic approaches that assume classic drift, migration, adaptation equilibrium that could take hundreds or thousands of years. However, these are also arguably the types of contexts most immediately relevant to understanding and managing human influences on evolution. To overcome these challenges and assumptions we often compare our genetic data with simulation models that account for contemporary time and spatial dynamics. Some recent and current projects on landscape genetics and genomics:
- Biogeography of Island Salamanders: Working with USGS, National Wildlife Refuge and National Park biologists we discovered that Maine’s coastal islands support numerous local populations of red-backed salamanders (Plethodon cinereus). Graduate student Nikko Shaidani is using population genetic approaches to determine whether and how these populations originated – post-glacial colonization or more recent transport by humans.
- Landscape genetics and genomics of vernal pool amphibians: As part of a larger NSF Coupled Natural Human Systems team, we are assessing how natural and anthropogenic landscape features, including urbanization patterns, influence the population structure and adaptations of vernal pool amphibians. These findings are part of Jared Homola’s dissertation and will be part of a larger modelling process that links vernal pool legislation, township and landowner actions, and vernal pool amphibian sustainability.
- Influence of past stocking on Maine’s wild trout: Maine is considered the last great stronghold of native brook trout (Salvelinus fontinalis) diversity in the US, but throughout the past century hatchery sources of trout were often stocked on top of wild populations leading to uncertainty about whether some of Maine’s trout populations are still representative of the local genetic heritage of the species. We are working with the Maine Department of Inland Fisheries and Wildlife to determine which wild trout populations might have been affected by past stocking and how much.

Environmental DNA (eDNA)
A very large portion of conservation in practice surrounds a problem akin to finding a few needles in hundreds or thousands of haystacks. The needles we are searching for in conservation are of course invasive or threatened species. Unlike the inanimate needles, where we might search at leisure with little negative consequences (hay fever?), these conservation needles are mobile, can actively avoid detection, and can change in abundance over short time frames. Moreover, these conservation needles are concealed in large and complex haystacks where sampling requires specialized equipment and is very inefficient. The cost of detecting and quantifying often small numbers of such species is immense, and that cost is not only monetary. The time and effort to ensure a high probability detection of a rare organism with traditional approaches is often beyond the capacity of most surveys. In some cases traditional sampling approaches like trapping, netting or angling also cause incidental injuries, mortality, or habitat destruction that would be highly undesirable, if not outright prohibited (e.g., take of an endangered species). It is an understatement that there is great need in conservation biology for a more sensitive, less costly and less damaging approach to detecting and quantifying rare organisms in difficult to sample habitats. Fortunately, evolution gives us that tool.
Reproductive isolation, mutation, genetic drift, and probably some selection, have in combination resulted in significant DNA sequence divergence among most species and even subspecies. This sequence variation has been analogized as a genetic barcode of sorts that can permit unique identification of taxa. When cells are shed by organisms as part of the normal maintenance of their bodies, reproduction, injury or death, that DNA persists for some period of time in the surrounding environment before it degrades into fragments too small for species identification. That shed cellular DNA is referred to as ‘Environmental DNA’, or ‘eDNA’, and it can be detected and quantified using modern genetic tools similar to those used in forensics.
In aquatic systems, eDNA often persists for weeks to months and aquatic organsim’s shed large amounts of it every day, in essence painting a giant DNA billboard with their name on it for us to detect with simple water samples. Obviously, there is more technology to it than that implies, and eDNA sampling brings with it its own sampling and error considerations for robust detection, but the relatively low cost, high sensitivity and low impact of eDNA approaches pretty much ensure they will become a widespread tool in conservation and natural resource management. A big part of this eDNA revolution requires establishing the DNA tools and sampling deigns appropriate to different habitats, sample types, regional species pools and inferences. We are currently undertaking this development process for many aquatic species of management concern in Maine and the Northeast U.S.
Some current development and applications of eDNA in our lab include:
- Native and Invasive Esox: In North America, the genus Esox includes Northern Pike, Muskellunge and a few Pickerel species. Non-native Northern Pike and Muskellunge are both major species of invasion concern in Maine. Redfin Pickerel are a state threatened species. Lauren Turinetti is developing eDNA tools for this group of fish and piloting a survey to quantify the extent of Northern Pike invasion in the Penobscot River system.
- Native and Invasive Centrarchids: The Centrarchidae is the family of fishes including bass and freshwater sunfishes. Several of these species are invasive concerns in Maine where their range is rapidly expanding via illegal introductions. Geneva York is developing eDNA detection tools for several of these species and piloting their use to verify confirmed and unconfirmed reports in multiple waterways.
- Salmonid extirpations and restoration: we are working with the State of Maine and NGOs to develop and apply eDNA to pressing questions in the management of endangered Atlantic Salmon, Brook Trout and other species. This includes potential verifications of suspected extirpations as well of potential access or colonization by these species into restored habitats.

Conservation and Ecology of Aquatic Species
While many of our lab activities center on evolution and its applications, we also have a long history of involvement in projects on other aspects of conservation of aquatic species, including threatened or endangered species as well as species of economic or cultural significance. Most of our work in this area has focused on salmonids (salmon, charr and trout) and sturgeons, but some new work is underway on vernal pool amphibians.
For my entire time in Maine I have served as an advisor on the genetic broodstock management for endangered Atlantic salmon. However, in addition my interests in the genetic and evolutionary management of the species, I have also been involved in a series of collaborative telemetry and passage studies with Joseph Zydlewski, and others, to characterize the anthropogenic migratory challenges faced by adult and juvenile salmon in the Penobscot River. That work represented an important set of pre-restoration insights in advance of the Penobscot River Restoration Project, one of the largest river restoration projects of its kind. Our new tracking studies have begun to reveal how dam removals and other changes the Penobscot system have changed migratory conditions for salmon.
The other Maine salmonid that has received ongoing attention in my lab is the Arctic charr (Salvelinus alpinus), or ‘blueback trout’ as it is commonly known in Maine. In addition to work demonstrating important ecotypic differences among the dozen populations of the species in Maine (the only state outside Alaska with native charr populations), the lab was involved in helping set guidelines for reclamation of Arctic charr populations threatened by smelt introductions. However, our longest-running Arctic charr project has involved a long-term mark-recapture, genetic and trait monitoring study of the spawning population of Arctic charr in Floods Pond. That dataset, based on collaborations with the Bangor Water District and Maine Department of Inland Fisheries and Wildlife, has the potential to offer unique insight into the ecology of this species at the very southern extent of its species range.
Brook trout (Salvelinus fontinalis) are gaining increasing attention in the lab. In addition to our population genetic work on trout stocking histories (see Landscape Genetics and Genomics section), we currently have a project underway with the Maine Department of Inland Fisheries and Wildlife and the Sea-Run Brook Trout Coalition to characterize patterns of marine migration (diadromy) in sea run brook trout in Maine based on elemental profiles in otoliths. Otolith analysis requires lethal sampling of fish, so as part of this project we are also attempting to develop a non-lethal stable isotope approach to confirm recent marine migration.
Ecology of Shortnose (Acipenser brevirostrum) and Atlantic Sturgeon (Acipenser oxyrinchus) have been a major research area of collaboration since 2005, when Gayle Zydlewski and I teamed up with a new graduate student to determine if any sturgeon inhabited the Penobscot River, a river with a long history of over-harvest, dams and pollution. We ended up capturing both species, which was remarkable in that shortnose had not been confirmed in the system in over 30 years and were thought to have likely been extirpated. In the ensuing years a great line-up of graduate students have characterized the abundance and movements of these sturgeon and revealed a previously unknown migratory life history for shortnose in the Gulf of Maine, where this typically estuary-resident species commonly migrates hundreds of ocean kilometers to other river system to feed or reproduce. Indeed, this life history is in many ways responsible for formation of a research network of sturgeon researchers throughout Gulf of Maine region.
Finally, in addition to our fish projects, we are now collaborating on projects surrounding the ecology of vernal pool amphibians and the complex ecological, social and economic processes at play in their conservation. This work involves collaborations with wildlife and wetland ecologists, as well as economists who specialize in modelling the socio-economic processes surrounding conservation behavior. Vernal pools are interesting and important systems to study coupled natural-human system dynamics because they are emblematic of small natural landscape features with disproportionately large ecological effects.
