Image Analysis – Software and Analysis Services
We are a full-service facility offering access to multiple image analysis software packages and providing expert consulting for analysis projects.
Our offerings include ImageJ for straightforward analysis as well as the Imaris for Core Facilities Package for high-content analysis and image processing.
ImageJ is a versatile platform developed by the NIH for all-access analysis needs.
Imaris is a user-friendly and extremely powerful package that excels in visually presenting 3D and 4D datasets. It boasts robust capabilities for conducting quantitative analyses on multi-dimensional images, and seamlessly manages a wide array of commercial fluorescence microscopy file formats.
Our Imaris workstation, built by Puget Systems, is fully equipped with all Imaris packages including ClearView deconvolution and Imaris Stitcher. It features a powerful AMD Ryzen Threadripper PRO 5975WX 32-core processor, 512 GB of RAM, and NVIDIA RTX 4000 Ada Generation graphics card, ensuring top-notch performance for advanced imaging data analysis.
Dr. Zhengxin Ma and Dr. Robert Wheeler are available for image analysis consultation. Please email Dr. Ma at zhengxin.ma@maine.edu.
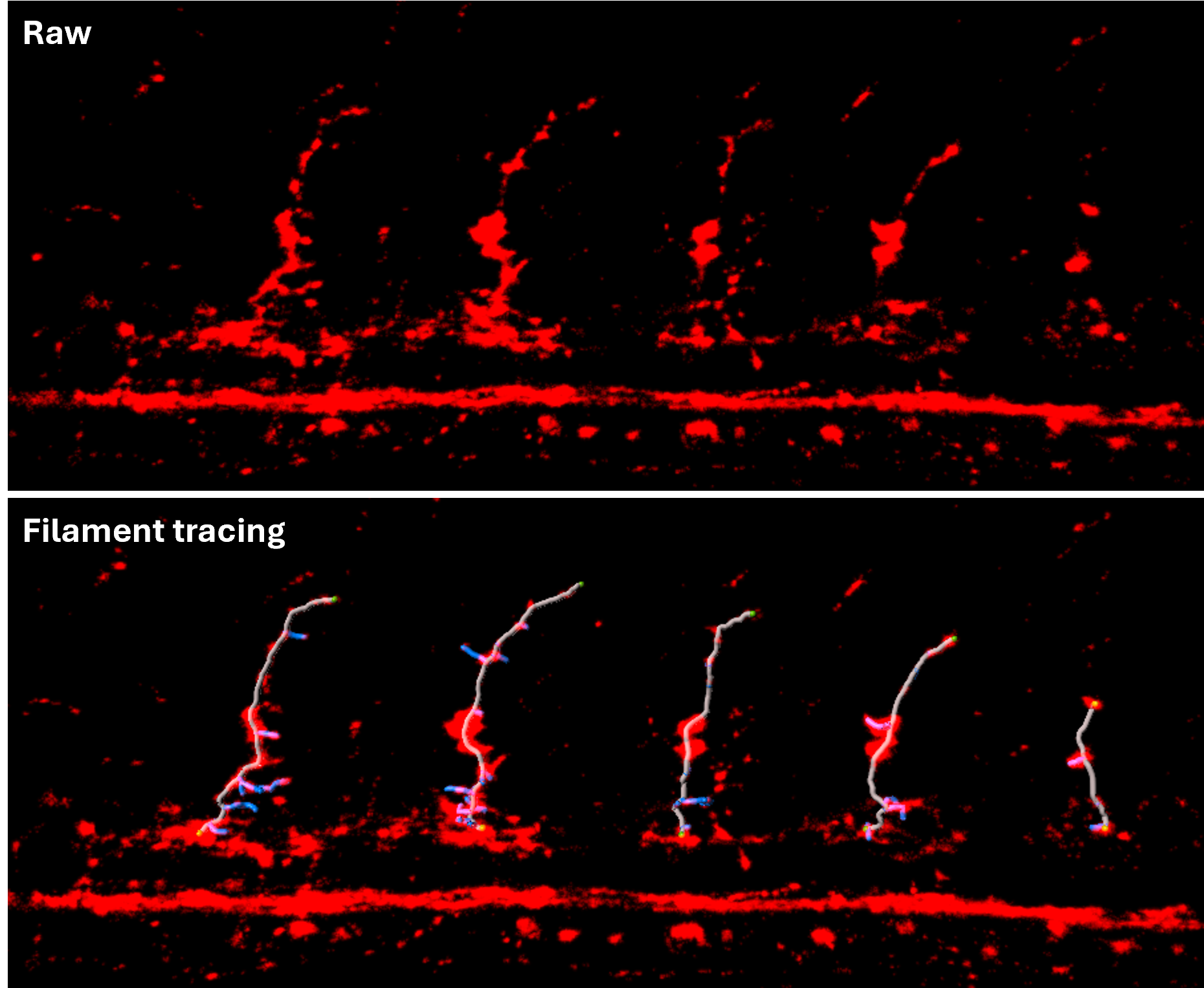
Filament tracing: Zebrafish primary motor neurons can be visualized and analyzed using the Filament Tool, which provides both manual and automatic tracing options. With this tool, you can measure each neuron’s length, count, branch numbers and more.
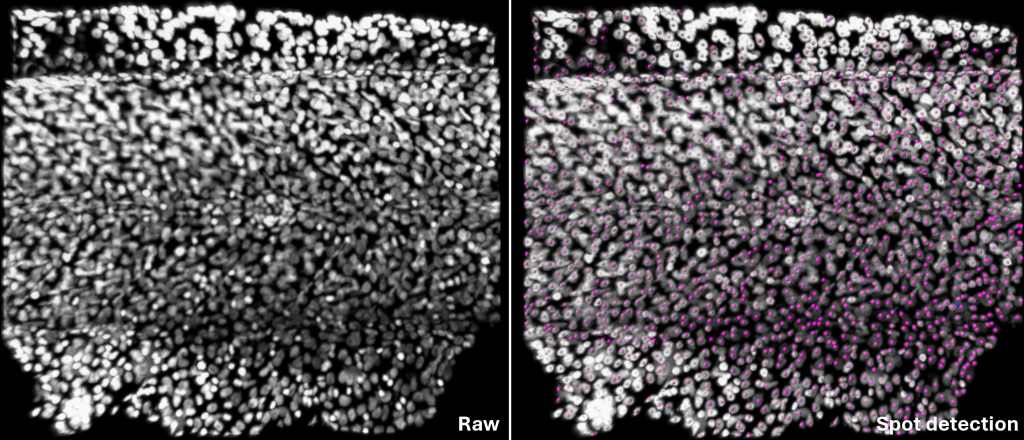
Spots: The nuclei are annotated and quantified using the Spots tool in Imaris. This tool automatically, also has the manual features, detects discrete points, enabling researchers to identify and count objects that are otherwise difficult to quantify.
Muscle fiber segmentation: Zebrafish muscle fibers can be annotated using Deep learning features within Surface Tool in Imaris. This tool supports 3D segmentation, analysis, and surface visualization, which helps researchers compare and measure muscle fiber structures.
Tracking macrophages: The Surface tool enables tracking of objects across time-lapse images, helping researchers follow changes in position, shape and movement over time. Once the objects are tracked, users can analyze their properties such as velocity, displacement, volume, and surface area to gain deeper insights into dynamic behaviors or morphological changes.
